





















MOLCAS manual: Next: 8.51 The Basis Set Libraries Up: 8. Programs Previous: 8.49 vibrot
|
| File | Contents |
| WFAH5 | All information that the WFA program needs is contained in this HDF5 file. The name can be adjusted with the H5FIle option. |
8.50.3.2 Output files
| File | Contents |
| WFAH5 | The orbital coefficients of NOs, NTOs, and NDOs are written to the same HDF5 file that is also used for input. |
| *atomic.om | These are input files for the external TheoDORE program. |
Extraction of the NOs, NTOs, and NDOs from the HDF5 file occurs with the external Molpy program. Call, e.g.:
penny molcas.rassi.h5 --wfaorbs molden
8.50.4 Input
The input for the WFA module is preceded by:
&WFA
8.50.4.1 Keywords
Basic Keywords:
| Keyword | Meaning |
| H5FIle | Specifies the name of the HDF5 file used for reading and writing
(e.g. $Project.scf.h5, $Project.rasscf.h5, $Project.rassi.h5).
You either have to use this option or rename the file of
interest to WFAH5.
|
| REFState | Index of the reference state for 1TDM and 1DDM analysis.
|
| WFALevel | Select how much output is produced (0-4, default: 3).
|
Advanced keywords for fine grain output options and debug information:
| Keyword | Meaning |
| MULLiken | Activate Mulliken population analysis.
|
| LOWDin | Activate Löwdin population analysis.
|
| NXO | Activate NO, NTO, and NDO analysis.
|
| EXCIton | Activate exciton and multipole analysis.
|
| CTNUmbers | Activate charge transfer number analysis and creation of *.om files.
|
| H5ORbitals | Print the NOs, NTOs, and/or NDOs to the HDF file.
|
| DEBUg | Print debug information.
|
| ADDInfo | Add info for molcas verify. |
8.50.4.2 Input example
* Analysis of SCF job
&SCF \\
&WFA
H5file = $Project.scf.h5 \\
* Analysis of RASSCF job
* Reduced output: only charge transfer numbers
&RASSCF \\
&WFA
H5file = $Project.rasscf.h5
wfalevel = 0
ctnumbers \\
* Analysis of RASSI job, use the TRD1 keyword
* Second state as reference
&RASSI
TRD1 \\
&WFA
H5file = $Project.rassi.h5
Refstate = 2
8.50.5 Output
8.50.5.1 State/difference density matrix analysis (SCF/RASSCF/RASSI)
RASSCF analysis for state 2 (3) A or
RASSI analysis for state R_2
| Descriptor | Explanation |
|---|---|
n_u |
Number of unpaired electrons
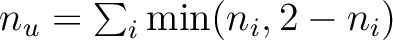 [185,179] [185,179] |
n_u,nl |
Number of unpaired electrons
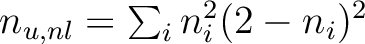 |
PR_NO |
NO participation ratio PRNO |
p_D and p_A |
Promotion number pD and pA |
PR_D and PR_A |
D/A participation ratio PRD and PRA |
<r_h> [Ang] |
Mean position of detachment density  [182] [182] |
<r_e> [Ang] |
Mean position of attachment density  |
|<r_e - r_h>| [Ang] |
Linear D/A distance
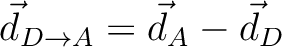 |
Hole size [Ang] |
RMS size of detachment density 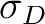 |
Electron size [Ang] |
RMS size of attachment density 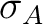 |
8.50.5.2 Transition density matrix analysis (RASSI)
RASSI analysis for transiton from state 1 to 2 (Tr_1-2)
| Output listing | Explanation |
|---|---|
Leading SVs |
Largest NTO occupation numbers |
Sum of SVs (Omega) |
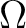 , Sum of NTO occupation numbers , Sum of NTO occupation numbers |
PR_NTO |
NTO participation ratio PRNTO [183] |
Entanglement entropy (S_HE) |
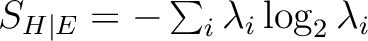 [186] [186] |
Nr of entangled states (Z_HE) |
ZHE=2SH|E |
Renormalized S_HE/Z_HE |
Replace
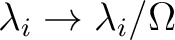 |
omega |
Norm of the 1TDM  , single-exc. character , single-exc. character |
<r_h> [Ang] |
Mean position of hole
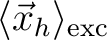 [182] [182] |
<r_e> [Ang] |
Mean position of electron
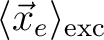 |
|<r_e - r_h>| [Ang] |
Linear e/h distance
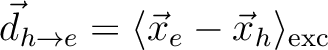 |
Hole size [Ang] |
RMS hole size:
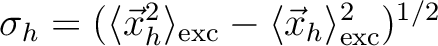 |
Electron size [Ang] |
RMS elec. size:
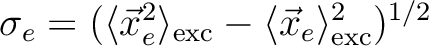 |
RMS electron-hole separation [Ang] |
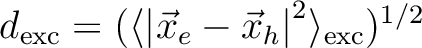 [181] [181] |
Covariance(r_h, r_e) [Ang^2] |
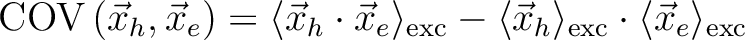 |
Correlation coefficient |
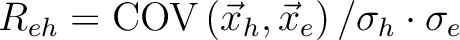 |
Next: 8.51 The Basis Set Libraries Up: 8. Programs Previous: 8.49 vibrot

